Since the emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Wuhan, China, in late December 2019, the virus has undergone several mutations that have led to the continuous emergence of variants.
With the implementation of monoclonal antibody therapeutics during the COVID19 pandemic, the selective pressures encountered by SARS-CoV-2 have been changed, increasing the probability that more resistant variants of the virus will develop and become transmissible into the larger population.
Some of the circulating variants are much more virulent than the ancestral strain and have made combatting the coronavirus disease 2019 (COVID-19) pandemic much harder. It is, therefore, essential to monitor the emergence and spread of new variants, as that will help in containing the transmission of the virus.
.jpg)
Background
The E484K mutation in the spike protein of SARS-CoV-2 has been found in a number of variants of concern, including B.1.1.7 (UK variant), B.1.351 (South Africa variant), and P.1 (Brazil variant). The spike protein is crucial for viral entry and is also the target for the majority of antibodies. Previous studies have shown that variants with the E484K mutation show reduced responses to vaccine-induced neutralizing antibodies.
The potential for monoclonal antibody treatments to select antibody-resistant mutants has been identified in previous research. However, the surveillance of close contacts or broader community of patients, post-administration, has not been reported.
A new study, published on the medRxiv* preprint server, suggested that selective pressures induced by the administration of monoclonal antibodies (Bamlanivimab) may require increased genomic surveillance to identify and control the spread of therapy-induced variants.
A New Study
The novelty of this study is that it is genomically supported and documents the onward transmission of a mutation of concern, which could have been selected post-monoclonal antibody therapy treatment of the index case.
The majority of single-agent monoclonal antibodies being used are a combination of products involving mixtures of antibodies, which should bring about a reduction in the selection of variants of concern. This should reduce the probability of selecting a resistance variant with mutations at two distinct epitopes and provide therapeutic durability in variants with a single mutation at one of the two designated epitopes that emerge.
Making antibodies cocktails from the list of approved agents is currently not permitted. However, scientists opine that this may provide for better treatment options. It would not only be beneficial for patients carrying the virus of known genotype but also substantially reduce the probability of the emergence of resistant variants.
Researchers also advocate for fast genotyping of the virus from the household members of patients who have received monoclonal antibody treatments. This could help in rigorous isolation of those carrying resistant variants and, thereby, prevent the onward spread of the same.
Previous research highlighted the many advantages of vaccination over monoclonal antibodies. A monoclonal antibody cannot cure individuals immediately, does not prevent transmission in the short term, does not guarantee against reinfection, performs poorly in populations experiencing exponential disease transmission, and diverts resources away from other areas where they are needed most during a crisis.
The current study adds one further cause for concern: widespread usage of monoclonal antibodies may select antibody-resistant strains that might also escape prior natural or vaccine-acquired immunity. To somewhat mitigate this problem, researchers suggest substantially improving surveillance in the context of monoclonal antibody utilization. However, the risks might still outweigh the potential benefits of the antibody treatment.
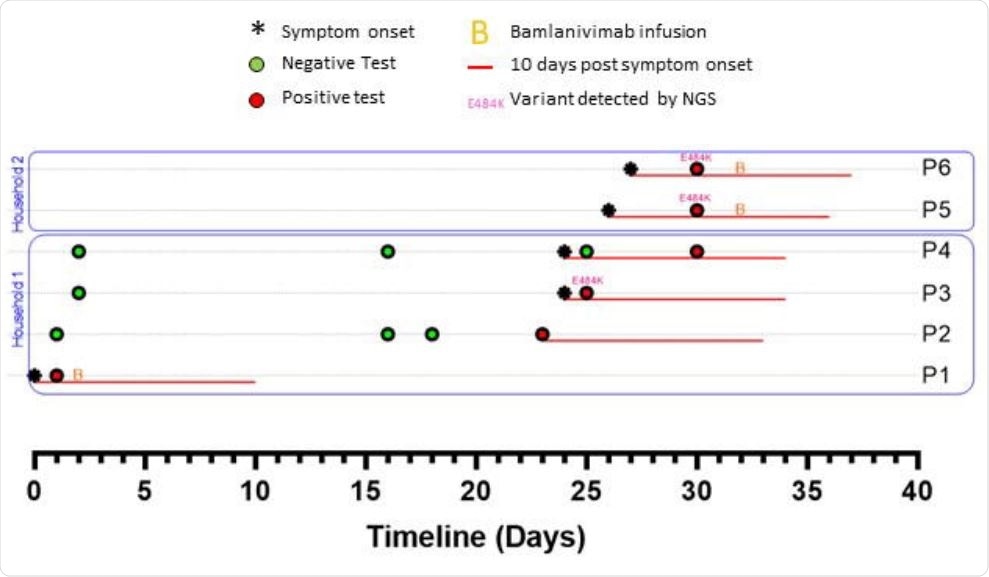
We should consider how the risks drive the broader epidemiology of the COVID-19 pandemic in the face of unrestrained global spread. Researchers stated that if this phenomenon of developing resistance was common, then we would observe more frequent examples of independent E484 mutations than what the current data show. Nevertheless, we should be mindful of these risks while conducting any cost-benefit analysis of deploying monoclonal antibody treatments at a large scale.
Even if the emerged resistance variants do not venture far from their point of origin, they may alter treatment outcomes locally. As an example, in the region where the authors of the current study are based, genome-level data would suggest a high degree of efficacy of Bamlanivimab/Etesevimab, despite being unavailable nationally. This would prevent local access to meaningful medication and the effect of this scenario is quite challenging to quantify.
Conclusion
The current study demonstrated the possibility of onward transmission of newly emerged antibody-resistant variants. These variants could emerge in patients with recent treatment history with single-agent monoclonal antibodies. These agents are widely being used, owing to which there is a possibility of reinforcing the existing genetic diversity of SARS-CoV-2. In the current study, scientists stated that if the use of monoclonal antibodies is to be further expanded, then there is a need for additional quick surveillance for mutations of concern.
*Important Notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Sabin, P, A., et al. (2021) Acquisition and onward transmission of a SARS-CoV-2 E484K variant among household contacts of a bamlanivimab-treated patient. medRxiv 2021.10.02.21264415; doi: https://doi.org/10.1101/2021.10.02.21264415, https://www.medrxiv.org/content/10.1101/2021.10.02.21264415v1
Posted in: Medical Research News | Disease/Infection News
Tags: Antibodies, Antibody, Coronavirus, Coronavirus Disease COVID-19, Efficacy, Epidemiology, Genetic, Genome, Genomic, Genotyping, immunity, Monoclonal Antibody, Mutation, Pandemic, Protein, Research, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Therapeutics, Vaccine, Virus

Written by
Dr. Priyom Bose
Priyom holds a Ph.D. in Plant Biology and Biotechnology from the University of Madras, India. She is an active researcher and an experienced science writer. Priyom has also co-authored several original research articles that have been published in reputed peer-reviewed journals. She is also an avid reader and an amateur photographer.
Source: Read Full Article
