Among the biggest challenges to containing the ongoing pandemic of coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is the emergence of mutations that allow the virus to evade host immune responses. Initially, changes in the SARS-CoV-2 genome were slow to emerge, due perhaps to the total lack of pre-existing immunity.
Intra-individual variation
A new preprint research paper posted to the bioRxiv* server reports the rapid emergence of variants within an infected individual. The evolution of the virus within an infected host is not excluded by the close similarity between the consensus sequences of single-person SARS-CoV-2, since the number of viral copies is at its peak soon after infection occurs and before the host develops adaptive immunity.
With the peak viral burden comes peak transmissibility, after which specific antibody and cellular responses may select for transmissible variants of the virus.
In the immunocompromised, with prolonged viral shedding, SARS-CoV-2 has been found to change its genomic sequence.
The occurrence of intra-individual variation is significant. One study has found an association between the number of genomic sites showing variation with disease severity at the time of sampling.
Overall, however, most individuals show stable consensus sequences over time, indicating that specific antiviral immune responses can continue to target the replicating viruses until the infection’s course is run.
That is, during the early phase of infection, SARS-CoV-2 has not been found to adapt in any single direction.
The issues with shotgun sequencing
A big issue with such studies is that the standard ‘shotgun’ methods used to sequence the virus from a given sample yield the consensus sample, hiding the multiple sequences found in a single individual due to the diverse viral quasispecies.
While shotgunning rapidly covers the whole length of the large viral genome in hundreds or thousands of samples simultaneously, thus detecting variation within a single individual worldwide, it has its limitations. The use of viral RNA amplification on fragments from multiple genomes, and shotgun sequencing of long regions in the form of multiple smaller fragments, prevents the recognition of genetic linkages and the correction of errors in individual haplotypes.
This means that variations within viral sequences within a single individual are reported only in the form of differences at genome positions that go beyond the normal background variation attributed to amplification and sequencing errors. This could swallow up true variations as well if found at low frequencies.
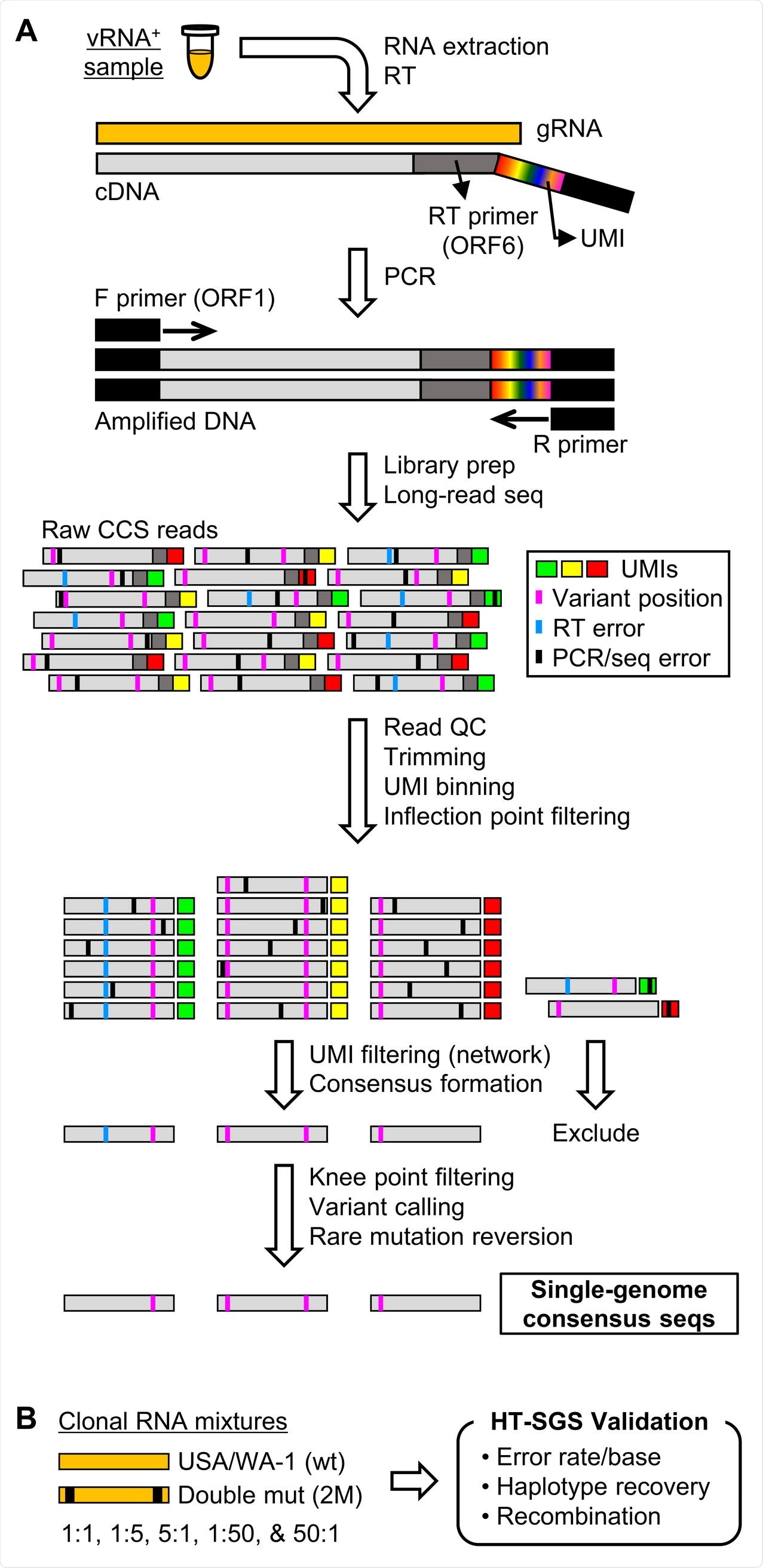
Changing the method around
The current study used a single-genome amplification and sequencing (SGS) approach to capture genetic variation within a single sample from an infected individual. The researchers introduced a high-throughput strategy (HT-SGS) whereby the surface protein gene region is subject to deep sequencing of long reads from large numbers of individual virus genomes.
Limited variation in host
The results show how SARS-CoV-2 gene variants emerged under selection pressure from host immune responses during the acute infection phase.
First, the investigators found extensive variations in the strains cultured in vitro, as a result of viral adaptation to the cultured cells, but only seven mutations found in the intra-individual variant haplotypes.
Among the seven participants in the study, the researchers found only one virus haplotype in 3 of them and 2-3 in the rest. The seven mutations were not found to form a structural signature that could be called an intra-individual variant haplotype.
Of these seven single nucleotide variants (SNVs), one SNV was found in the downstream of the spike gene, four in non-structural open reading frames (ORF) 3 and ORF6, and two synonymous SNVs. In fact, the SARS-CoV-2 sequences from these seven individuals were relatively homogeneous compared to those from the cultured virus.
This appeared to mirror the slow emergence of genetic variations in the virus during the early part of the pandemic.
Selection for neutralization epitope
However, when spike-antibody binding was analyzed in a single individual over time, they found that the total serum binding to the spike protein increased six-fold over days 12-16. Specific antibody binding was found at the N-terminal domain (NTD), receptor-binding domain (RBD), and S2 subunit.
The binding response widened in range over days 16-19, not being inhibited by any of a panel of monoclonal antibodies. This suggested that the genetically homogeneous samples of the cross-sectional phase of this study came from a period before circulating antibody responses to the viral spike protein had reached their peak.
The viral RNA burden showed a marked, though not steady, decline between days 9 and 17 in two phases. The first decline occurred over four orders of magnitude, between days 9 and 13, but then increased on day 15. It then fell again on day 17.
Interestingly, four minor haplotype variants emerged on day 15. All together carried three separate non-synonymous mutations affecting the same NTD epitope.
This site is neutralized by 4A8, and is a site often mutated in chronic SARS-CoV-2 infections, as well as in regions with recurrent deletions.
Before and after
Before these mutations emerged, serum antibodies from the same patient were observed to recognize the 4A8 site primarily. This suggests that as anti-spike NTD antibody titers rise in serum, multiple mutations emerge independently to evade the recognition of this site by these antibodies, leading to a slight interruption of viral clearance from the host.
In fact, it is possible that SARS-CoV-2 spike variants were selected for mutational immune escape by increasing antibody responses during acute infection. The lack of more variants of the virus genome was thus also potentially due to this factor.
Overlooked trend
This trend may have been overlooked, firstly because new mutations were being identified and followed on a global scale, focusing on cross-sectional studies and not on longitudinal follow-up. Secondly, the RNA obtained from early samples, when the viral RNA is at its peak, is of high quality, which could have led to this period of the infection being over-studied, at the expense of the viral genome later on, after adaptive immunity kicked in.
Thirdly, the current study used a method designed to analyze single virus genomes with multiple error correction layers, thus minimizing artefactual errors while picking up true genomic variations.
“This allowed groups of important virus variants to be detected even though each variant individually accounted for a small proportion of all sequences in each sample.”
Lastly, the patient in question had a history of stem cell transplantation, implying that immunosuppression might have played a role in the emergence of variants due to the higher rate of replication. The patient was not on immunomodulatory medication when diagnosed with COVID-19. However, viral loads were commensurate with those expected from earlier studies in immunocompetent individuals.
Immune response broadens to overcome evasion
Interestingly, the variants that emerged were replaced by wild-type sequences, probably due to the broadening of the immune response, which overcame the 4A8 site evasion. Thus, polyclonal responses appear to limit the viral escape from immunity, as shown by the absence of RBD variants, particularly in indel-intolerant genome regions.
Conversely, the newly emergent UK, South African and Brazil variants show that immune escape does occur. The extent to which viral clearance occurs is possibly linked to COVID-19 severity.
What are the implications?
The researchers concluded that this virus could rapidly adapt its genome to evade neutralizing antibodies, which could lead in part to delayed clearance of the virus during the acute phase of SARS-CoV-2 infection.
“It will be important to examine whether this reflects a “tipping point” in early infection at which SARS-CoV-2 genetic diversity can occasionally allow sustained replication through the evasion of immune recognition. Immunity induced by prior infection, vaccination, or passive immunization could reduce the potential for escape by controlling initial levels of virus replication quickly.”
Early antiviral or monoclonal antibody cocktail therapy could thus help overcome the infection much more effectively than treating with a single drug later on in the disease.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Ko, S. H. et al. (2021). High-Throughput, Single-Copy Sequencing Reveals SARS-CoV-2 Spike Variants Coincident with Mounting Humoral Immunity during Acute COVID-19. bioRxiv preprint. doi: https://doi.org/10.1101/2021.02.21.432184. https://www.biorxiv.org/content/10.1101/2021.02.21.432184v1
Posted in: Medical Research News | Disease/Infection News
Tags: Antibodies, Antibody, Cell, Chronic, Coronavirus, Coronavirus Disease COVID-19, Evolution, Gene, Genes, Genetic, Genome, Genomic, Immune Response, Immunization, Immunomodulatory, Immunosuppression, in vitro, Knee, Molecule, Monoclonal Antibody, Nucleotide, Pandemic, Protein, Receptor, Research, Respiratory, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Shotgun Sequencing, Spike Protein, Syndrome, Virus

Written by
Dr. Liji Thomas
Dr. Liji Thomas is an OB-GYN, who graduated from the Government Medical College, University of Calicut, Kerala, in 2001. Liji practiced as a full-time consultant in obstetrics/gynecology in a private hospital for a few years following her graduation. She has counseled hundreds of patients facing issues from pregnancy-related problems and infertility, and has been in charge of over 2,000 deliveries, striving always to achieve a normal delivery rather than operative.
Source: Read Full Article
