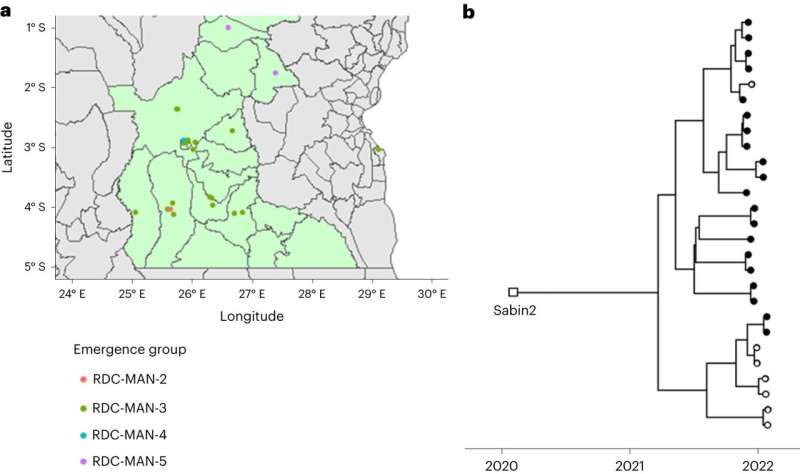
A new study, “Sensitive poliovirus detection using nested PCR and nanopore sequencing: a prospective validation study,” published 17 August 2023 in Nature Microbiology, proves that using DDNS to detect polio outbreaks can save public health authorities crucial time and money.
This research was jointly conducted by researchers at the Institut National de Recherche Biomédicale in Kinshasa who implemented DDNS in the Democratic Republic of the Congo (DRC) for the detection of polio outbreaks in collaboration with the MHRA, Imperial College London, the University of Edinburgh and various laboratories of the World Health Organization (WHO) Global Polio Laboratory Network (GPLN).
This is the first time that this type of scientific technique has been used to detect polio. Similar techniques have previously been used to detect COVID-19, Ebola, measles and monkeypox.
By enabling samples to be tested in the country where the outbreak originated rather than being sent to specialist laboratories abroad, the costs and delays of transport and testing can be reduced from an average of 42 days to an average of 19 days.

Currently, stool samples from countries with active polio outbreaks such as the DRC must be shipped around the world for lengthy, complex laboratory tests to confirm a polio case. Faster detection of polio in the regions where outbreaks still occur allows for a faster response by authorities through targeted, localized vaccination campaigns, minimizing the opportunity for the virus to spread.
Javier Martin, principal scientist in virology at the MHRA said, “We are standing at a delicate and pivotal moment for the eradication of polio. While vaccination programs have seen polio disappear in many countries, the delayed detection of outbreaks poses a major threat to those efforts.
“By implementing detection methods such as DDNS, we can identify where outbreaks are and which polio strain is present much more quickly, allowing us to act at the earliest opportunity. This is the result of years of work, collaborating with our partners. Together, we will continue to build on this research and support countries at risk of outbreaks to implement DDNS testing to help make polio a disease of the past.”
This research showed that DDNS tests done locally in the DRC over a six-month period were an average of 23 days faster than the standard method, with more than 99% accuracy.
Researchers also tested this technique in the U.K. and detected poliovirus in London in 2022, leading to the recent drive to ensure children under the age of 12 are vaccinated through the London polio catch-up campaign 2023.
Professor Placide Mbala-Kingebeni, Medical Doctor and Virologist at the Institut National de Recherche Biomédicale said, “This is the perfect example of collaboration, where combining and sharing knowledge together with all our partners has supported the vital work of the INRB in the DRC where poliomyelitis remains a serious public health problem.
“Collaboration and training with our partners has empowered the local team not only to master and confidently carry out this new technique but also to transfer the knowledge and skills to other African countries where poliovirus outbreaks are reported regularly.”
Dr. Alex Shaw, research fellow in the School of Public Health at Imperial College London said, “This method allows the rapid confirmation of polio strains, facilitating swifter vaccine responses that can reduce the number of polio cases stemming from an outbreak. Development and validation of the method has been the result of fruitful collaboration between a consortium of many partners.
“As a consortium we look forward to the training of additional national laboratories in this method, with prior trainees, including members of INRB, now taking on the role of trainers. The sequencing technology used in this method is easily adapted for the detection and typing of other organisms. This rollout will therefore provide a foundation of skills and experience that can be redirected to the genomic surveillance of other pathogens as needed.”
Polio is an infectious disease caused by the poliovirus, most commonly transmitted through contact with infected feces via contaminated food and water.
While many people may never show symptoms, in extreme cases, especially for babies and children under the age of five, polio can lead to permanent paralysis or death.
The WHO has identified delays in detection as one of the major challenges facing their Polio eradication strategy 2022–2026.
While faster detection methods such as DDNS cannot eradicate polio on their own, they play an essential part in managing outbreaks.
Scientists at the MHRA will continue to support the testing and validation of DDNS as a polio detection technique and training WHO laboratories around the world in how to use it.
More information:
Alexander G. Shaw et al, Sensitive poliovirus detection using nested PCR and nanopore sequencing: a prospective validation study, Nature Microbiology (2023). DOI: 10.1038/s41564-023-01453-4
Journal information:
Nature Microbiology
Source: Read Full Article