Research led by the Sun Yat-sen University Cancer Center in China has highlighted genetic risks for nasopharyngeal carcinoma. In a paper titled “High-throughput identification of regulatory elements and functional assays to uncover susceptibility genes for nasopharyngeal carcinoma” published in The American Journal of Human Genetics, the international team applied multiple genomic approaches in identifying genes critical to the development of nasopharyngeal carcinoma.
Nasopharyngeal carcinoma (NPC) is an aggressive malignancy that originates in the upper part of the throat behind the nose and is associated with Epstein-Barr virus infection, a member of the herpes virus family that causes mononucleosis or “mono.” Epstein-Barr infections are global and so common that most humans get infected with the virus at some point.
The Epstein-Barr virus has been risk associated with NPC, with a heightened risk factor for those of Chinese descent. The incidence of NPC is highest in east and southeast Asia, accounting for 77% of new NPC cases worldwide, implying a robust genetic risk aspect to the disease development.
Previous work by the team and other research institutions had identified many genetic associations with NPC. The net cast by these previous studies was wide, leaving specific disease mechanisms hidden within the larger web of interactions.
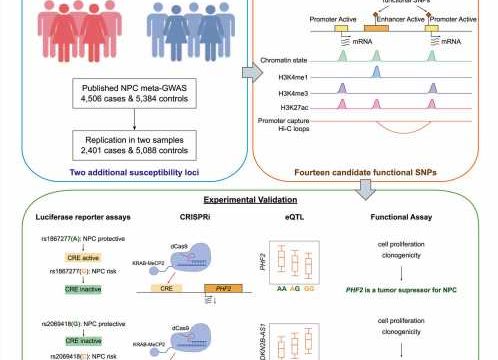
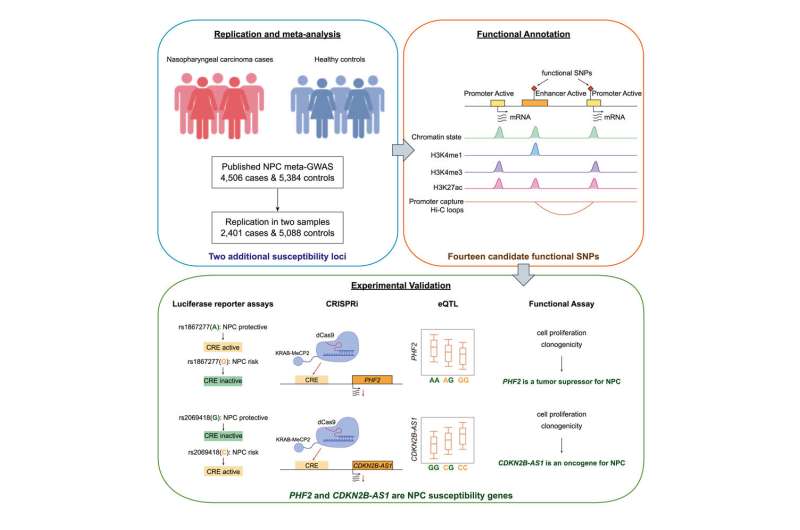
The current study performed genetic associations by combining 6,907 cases and 10,472 controls to identify additional NPC susceptibility loci. The findings suggest that genes PHF2 and CDKN2B-AS1 may play essential roles in NPC development and highlight the significance of the DNA damage repair pathway.
The study integrated epidemiological studies with genomic profiling and experiments to provide deeper evidence for the association between single nucleotide polymorphisms (SNPs) and NPC susceptibility. They found that cis-regulatory sequences exhibit different regulatory activity when harboring risk or protective alleles of SNPs.
Cis-regulatory sequences, such as enhancers and promoters, can control physiology by regulating access to genes affecting when and with what level of intensity the genes are expressed.
The risk alleles identified reduced the enhancer activity of a regulatory element, inhibiting the expression of the target gene PHF2. Reduction of PHF2 promoted cell proliferation and clonogenicity, suggesting its role as a tumor suppressor in NPC.
The risk alleles in the same locus increased the enhancer activity of another regulatory element, leading to higher expression of CDKN2B-AS1. Overexpression of CDKN2B-AS1 promoted cell proliferation and clonogenicity, indicating its oncogenic role in NPC.
The researchers suggest that the SNPs identified could be used as biomarkers for NPC risk prediction and that further investigation is warranted to elucidate the molecular mechanism of the identified susceptibility genes.
More information:
Tong-Min Wang et al, High-throughput identification of regulatory elements and functional assays to uncover susceptibility genes for nasopharyngeal carcinoma, The American Journal of Human Genetics (2023). DOI: 10.1016/j.ajhg.2023.06.003
Journal information:
American Journal of Human Genetics
Source: Read Full Article