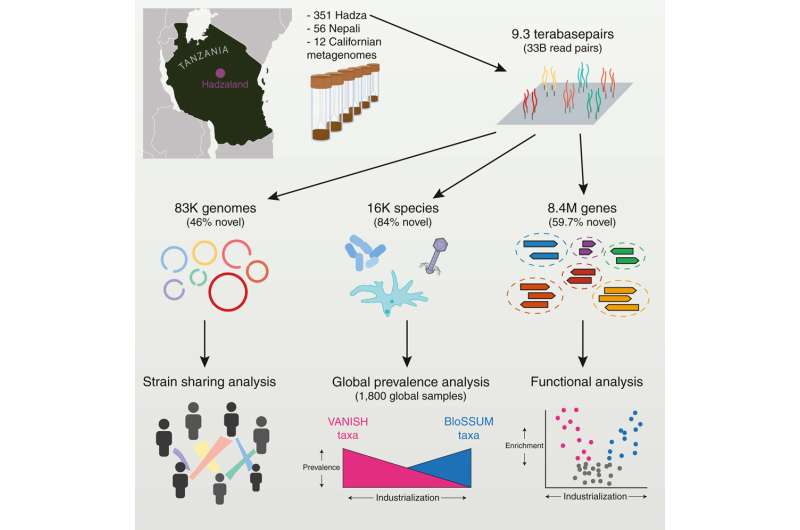
A genomic study led by researchers at Stanford University, California, has performed ultra-deep metagenomic sequencing on 351 fecal samples from the Hadza hunter-gatherers of Tanzania. In their paper, “Ultra-deep sequencing of Hadza hunter-gatherers recovers vanishing gut microbes,” published in Cell, the team details their collection and identification of the most extensive set of gut microbiome sequencing data from a hunter-gatherer population.
Microbiome composition can vary significantly worldwide, depending on local diet and environmental contact. Microbiome studies are heavily biased toward Western industrialized populations with low microbiome diversity. The low diversity is thought to be caused by intake of highly processed foods, more frequent antibiotic use, cesarean section rates, sanitation levels and reduced physical contact with animals and soil.
This causal association is supported by looking at non-industrialized populations, including hunter-gatherers, with extremely high microbiome diversity. It can also be observed in the altered microbiome of immigrants from non-industrialized areas to industrialized Western cultures as a transition toward lower microbiome diversity.
Previous analysis of ancient human coprolites shows that ancient microbiomes more closely resemble the high diversity of the modern non-industrialized microbiome than the industrialized one.
These human-associated microbial lineages have been passed across hominid generations over deep evolutionary time, and knowing that gut microbiota plays essential roles in human health, suggests that the now missing aspects of the industrialized microbiome mean a loss of functions that these microbes provided.
The Hadza people in the current study live in the central Rift Valley of Tanzania in camps of approximately five to 30 people. Individuals move between camp locations about every four months. They primarily drink from water springs and streams and have a diet that includes foraged tubers, berries, honey, and hunted animals.
Metagenomic sequencing was conducted on stool samples collected from 167 Hadza individuals. Over half (59.7%) of the 8.4 million protein families found in Hadza gut microbes are absent from the Unified Human Gastrointestinal Protein database. Bacterial and viral species found in the microbiomes increase the global catalog of known microbiota by around 24%.
For a local comparison, the team compared the findings to adult Californians and found that while the average Hadza adult gut microbiome contains 730 species, there were only 277 species in the Californian gut microbiome.
The authors state, “An important challenge is to characterize the impact of these microbes on human physiology and determine in which contexts the absence or presence of species and functions are beneficial or detrimental to human health.”
It will be fascinating to know the impacts of the lack of diversity on Western metabolisms or what diseases, if any, the Hadza populations are avoiding by having this robust diversity. It is unlikely that the missing microbiota from the industrialized populations was there without a reason, though it could simply be that the reasons no longer apply.
Thanks to the current effort, future researchers have many new mysteries to solve and an ultra-deep sequenced data set to guide their questioning.
More information:
Matthew M. Carter et al, Ultra-deep sequencing of Hadza hunter-gatherers recovers vanishing gut microbes, Cell (2023). DOI: 10.1016/j.cell.2023.05.046
Journal information:
Cell
Source: Read Full Article